此处将为大家介绍关于Pythonnumpy模块-arctanh()实例源码的详细内容,并且为您解答有关pythonnumpyargmax的相关问题,此外,我们还将为您介绍关于Jupyter中的Nump
此处将为大家介绍关于Python numpy 模块-arctanh() 实例源码的详细内容,并且为您解答有关python numpy argmax的相关问题,此外,我们还将为您介绍关于Jupyter 中的 Numpy 在打印时出错(Python 版本 3.8.8):TypeError: 'numpy.ndarray' object is not callable、numpy.random.random & numpy.ndarray.astype & numpy.arange、numpy.ravel()/numpy.flatten()/numpy.squeeze()、Numpy:数组创建 numpy.arrray() , numpy.arange()、np.linspace ()、数组基本属性的有用信息。
本文目录一览:- Python numpy 模块-arctanh() 实例源码(python numpy argmax)
- Jupyter 中的 Numpy 在打印时出错(Python 版本 3.8.8):TypeError: 'numpy.ndarray' object is not callable
- numpy.random.random & numpy.ndarray.astype & numpy.arange
- numpy.ravel()/numpy.flatten()/numpy.squeeze()
- Numpy:数组创建 numpy.arrray() , numpy.arange()、np.linspace ()、数组基本属性

Python numpy 模块-arctanh() 实例源码(python numpy argmax)
Python numpy 模块,arctanh() 实例源码
我们从Python开源项目中,提取了以下44个代码示例,用于说明如何使用numpy.arctanh()。
- def test_branch_cuts(self):
- # check branch cuts and continuity on them
- yield _check_branch_cut, np.log, -0.5, 1j, 1, -1, True
- yield _check_branch_cut, np.log2, -0.5, np.log10, -0.5, np.log1p, -1.5, np.sqrt, True
- yield _check_branch_cut, np.arcsin, [ -2, 2], [1j, 1j], np.arccos, np.arctan, [0-2j, 2j], [1, 1], np.arcsinh, 2j], [1, 1], np.arccosh, [ -1, 0.5], [1j, 1j], np.arctanh, 2], True
- # check against bogus branch cuts: assert continuity between quadrants
- yield _check_branch_cut, [ 1, 1
- yield _check_branch_cut, 2], 1
- yield _check_branch_cut, 2, 0], 1], 2j, 1, 1
- def test_branch_cuts_complex64(self):
- # check branch cuts and continuity on them
- yield _check_branch_cut, True, np.complex64
- yield _check_branch_cut, np.complex64
- yield _check_branch_cut, np.complex64
- # check against bogus branch cuts: assert continuity between quadrants
- yield _check_branch_cut, False, np.complex64
- def test_against_cmath(self):
- import cmath
- points = [-1-1j, -1+1j, +1-1j, +1+1j]
- name_map = {''arcsin'': ''asin'', ''arccos'': ''acos'', ''arctan'': ''atan'',
- ''arcsinh'': ''asinh'', ''arccosh'': ''acosh'', ''arctanh'': ''atanh''}
- atol = 4*np.finfo(np.complex).eps
- for func in self.funcs:
- fname = func.__name__.split(''.'')[-1]
- cname = name_map.get(fname, fname)
- try:
- cfunc = getattr(cmath, cname)
- except AttributeError:
- continue
- for p in points:
- a = complex(func(np.complex_(p)))
- b = cfunc(p)
- assert_(abs(a - b) < atol, "%s %s: %s; cmath: %s" % (fname, p, a, b))
- def test_energy_conservation_sech2disk_manyparticles():
- # Test that energy is conserved for a self-gravitating disk
- N= 101
- totmass= 1.
- sigma= 1.
- zh= 2.*sigma**2./totmass
- x= numpy.arctanh(2.*numpy.random.uniform(size=N)-1)*zh
- v= numpy.random.normal(size=N)*sigma
- v-= numpy.mean(v) # stabilize
- m= numpy.ones_like(x)/N*(1.+0.1*(2.*numpy.random.uniform(size=N)-1))
- g= wendy.nbody(x,v,m,0.05)
- E= wendy.energy(x,m)
- cnt= 0
- while cnt < 100:
- tx,tv= next(g)
- assert numpy.fabs(wendy.energy(tx,tv,m)-E) < 10.**-10., "Energy not conserved during simple N-body integration"
- cnt+= 1
- return None
- def test_energy_conservation_sech2disk_manyparticles():
- # Test that energy is conserved for a self-gravitating disk
- N= 101
- totmass= 1.
- sigma= 1.
- zh= 2.*sigma**2./totmass
- x= numpy.arctanh(2.*numpy.random.uniform(size=N)-1)*zh
- v= numpy.random.normal(size=N)*sigma
- v-= numpy.mean(v) # stabilize
- m= numpy.ones_like(x)/N*(1.+0.1*(2.*numpy.random.uniform(size=N)-1))
- omega= 1.1
- g= wendy.nbody(x,0.05,omega=omega)
- E= wendy.energy(x,omega=omega)
- cnt= 0
- while cnt < 100:
- tx,omega=omega)-E) < 10.**-10., "Energy not conserved during simple N-body integration with external harmonic potential"
- cnt+= 1
- return None
- def test_energy_conservation_sech2disk_manyparticles():
- # Test that energy is conserved for a self-gravitating disk
- N= 101
- totmass= 1.
- sigma= 1.
- zh= 2.*sigma**2./totmass
- x= numpy.arctanh(2.*numpy.random.uniform(size=N)-1)*zh
- v= numpy.random.normal(size=N)*sigma
- v-= numpy.mean(v) # stabilize
- m= numpy.ones_like(x)/N*(1.+0.1*(2.*numpy.random.uniform(size=N)-1))
- g= wendy.nbody(x,approx=True,nleap=1000)
- E= wendy.energy(x,m)-E)/E < 10.**-6., "Energy not conserved during approximate N-body integration"
- cnt+= 1
- return None
- def test_notracermasses():
- # approx should work with tracer sheets
- # Test that energy is conserved for a self-gravitating disk
- N= 101
- totmass= 1.
- sigma= 1.
- zh= 2.*sigma**2./totmass
- x= numpy.arctanh(2.*numpy.random.uniform(size=N)-1)*zh
- v= numpy.random.normal(size=N)*sigma
- v-= numpy.mean(v) # stabilize
- m= numpy.ones_like(x)/N*(1.+0.1*(2.*numpy.random.uniform(size=N)-1))
- m[N//2:]= 0.
- m*= 2.
- g= wendy.nbody(x, "Energy not conserved during approximate N-body integration with some tracer particles"
- cnt+= 1
- return None
- def test_energy_conservation_sech2disk_manyparticles():
- # Test that energy is conserved for a self-gravitating disk
- N= 101
- totmass= 1.
- sigma= 1.
- zh= 2.*sigma**2./totmass
- x= numpy.arctanh(2.*numpy.random.uniform(size=N)-1)*zh
- v= numpy.random.normal(size=N)*sigma
- v-= numpy.mean(v) # stabilize
- m= numpy.ones_like(x)/N*(1.+0.1*(2.*numpy.random.uniform(size=N)-1))
- omega= 1.1
- g= wendy.nbody(x,omega=omega,omega=omega)-E)/E < 10.**-6., "Energy not conserved during approximate N-body integration with external harmonic potential"
- cnt+= 1
- return None
- def test_againstexact_sech2disk_manyparticles():
- # Test that the exact N-body and the approximate N-body agree
- N= 101
- totmass= 1.
- sigma= 1.
- zh= 2.*sigma**2./totmass
- x= numpy.arctanh(2.*numpy.random.uniform(size=N)-1)*zh
- v= numpy.random.normal(size=N)*sigma
- v-= numpy.mean(v) # stabilize
- m= numpy.ones_like(x)/N*(1.+0.1*(2.*numpy.random.uniform(size=N)-1))
- omega= 1.1
- g= wendy.nbody(x,nleap=2000,omega=omega)
- ge= wendy.nbody(x,tv= next(g)
- txe,tve= next(ge)
- assert numpy.all(numpy.fabs(tx-txe) < 10.**-5.), "Exact and approximate N-body give different positions"
- assert numpy.all(numpy.fabs(tv-tve) < 10.**-5.), "Exact and approximate N-body give different positions"
- cnt+= 1
- return None
- def test_branch_cuts(self):
- # check branch cuts and continuity on them
- yield _check_branch_cut, 1
- def test_branch_cuts_complex64(self):
- # check branch cuts and continuity on them
- yield _check_branch_cut, np.complex64
- def test_against_cmath(self):
- import cmath
- points = [-1-1j, b))
- def test_branch_cuts(self):
- # check branch cuts and continuity on them
- yield _check_branch_cut, 1
- def test_branch_cuts_complex64(self):
- # check branch cuts and continuity on them
- yield _check_branch_cut, np.complex64
- def test_against_cmath(self):
- import cmath
- points = [-1-1j, b))
- def test_branch_cuts(self):
- # check branch cuts and continuity on them
- yield _check_branch_cut, 1
- def test_branch_cuts_complex64(self):
- # check branch cuts and continuity on them
- yield _check_branch_cut, np.complex64
- def test_against_cmath(self):
- import cmath
- points = [-1-1j, b))
- def test_branch_cuts(self):
- # check branch cuts and continuity on them
- yield _check_branch_cut, 1
- def test_branch_cuts_complex64(self):
- # check branch cuts and continuity on them
- yield _check_branch_cut, np.complex64
- def test_against_cmath(self):
- import cmath
- points = [-1-1j, b))
- def test_branch_cuts(self):
- # check branch cuts and continuity on them
- yield _check_branch_cut, 1
- def test_branch_cuts_complex64(self):
- # check branch cuts and continuity on them
- yield _check_branch_cut, np.complex64
- def test_against_cmath(self):
- import cmath
- points = [-1-1j, b))
- def get_net_vectors(subject_list, kind, atlas_name="aal"):
- """
- subject_list : the subject short IDs list
- kind : the kind of connectivity to be used,e.g. lasso,partial correlation,correlation
- atlas_name : name of the atlas used
- returns:
- matrix : matrix of connectivity vectors (num_subjects x num_connections)
- """
- # This is an alternative implementation
- networks = load_all_networks(subject_list, atlas_name=atlas_name)
- # Get Fisher transformed matrices
- norm_networks = [np.arctanh(mat) for mat in networks]
- # Get upper diagonal indices
- idx = np.triu_indices_from(norm_networks[0], 1)
- # Get vectorised matrices
- vec_networks = [mat[idx] for mat in norm_networks]
- # Each subject should be a row of the matrix
- matrix = np.vstack(vec_networks)
- return matrix
- def test_branch_cuts(self):
- # check branch cuts and continuity on them
- yield _check_branch_cut, 1
- def test_branch_cuts_complex64(self):
- # check branch cuts and continuity on them
- yield _check_branch_cut, np.complex64
- def test_against_cmath(self):
- import cmath
- points = [-1-1j, b))
- def transformparameterndarray(parameterndarray, includejumps):
- parameterndarray = npu.tondim1(parameterndarray)
- res = [
- parameterndarray[0], # meanlogvar
- 2. * np.arctanh(parameterndarray[1]), # persistence
- np.log(parameterndarray[2] * parameterndarray[2]), # voloflogvar
- 2. * np.arctanh(parameterndarray[3]) # cor
- ]
- if includejumps:
- res.append(np.arctanh(2*parameterndarray[4] - 1)) # jumpintensity
- res.append(np.log(parameterndarray[5] * parameterndarray[5])) # jumpvol
- return np.array(res)
- def g_inv(x, t=4):
- """Inverse of g transform."""
- xp = np.clip(x, -t, t)
- diff = np.arctanh(np.clip(x - xp, -1 + 1e-10, 1 - 1e-10))
- return xp + diff
- def initialize(self, z0):
- z = self.opt_model[2]
- z.set_value(floatX(np.arctanh(z0)))
- def invert_bfgs(gen_model, invert_model, ftr_model, im, z_predict=None, npx=64):
- _f, z = invert_model
- nz = gen_model.nz
- if z_predict is None:
- z_predict = np_rng.uniform(-1., 1., size=(1, nz))
- else:
- z_predict = floatX(z_predict)
- z_predict = np.arctanh(z_predict)
- im_t = gen_model.transform(im)
- ftr = ftr_model(im_t)
- prob = optimize.minimize(f_bfgs, z_predict, args=(_f, im_t, ftr),
- tol=1e-6, jac=True, method=''L-BFGS-B'', options={''maxiter'':200})
- print(''n_iters = %3d,f = %.3f'' % (prob.nit, prob.fun))
- z_opt = prob.x
- z_opt_n = floatX(z_opt[np.newaxis, :])
- [f_opt, g, gx] = _f(z_opt_n, ftr)
- gx = gen_model.inverse_transform(gx, npx=npx)
- z_opt = np.tanh(z_opt)
- return gx, z_opt,f_opt
- def test_time():
- # Just run the timer...
- N= 101
- totmass= 1.
- sigma= 1.
- zh= 2.*sigma**2./totmass
- x= numpy.arctanh(2.*numpy.random.uniform(size=N)-1)*zh
- v= numpy.random.normal(size=N)*sigma
- v-= numpy.mean(v) # stabilize
- m= numpy.ones_like(x)/N*(1.+0.1*(2.*numpy.random.uniform(size=N)-1))
- g= wendy.nbody(x,nleap=1000,full_output=True)
- tx, time_elapsed= next(g)
- assert time_elapsed < 1., ''More than 1 second elapsed for simple problem''
- return None
- def itransform_define(transform):
- """
- This function links the user''s choice of transformation with its inverse
- """
- if transform == ''tanh'':
- return np.arctanh
- elif transform == ''exp'':
- return np.log
- elif transform == ''logit'':
- return Family.logit
- elif transform is None:
- return np.array
- else:
- return None
- def itransform_name_define(transform):
- """
- This function is used for model results table,displaying any transformations performed
- """
- if transform == ''tanh'':
- return ''arctanh''
- elif transform == ''exp'':
- return ''log''
- elif transform == ''logit'':
- return ''ilogit''
- elif transform is None:
- return ''''
- else:
- return None
- def atanh(v):
- return v.__class__(numpy.arctanh(v))
- def inv_clipping_sigma(x, max_in):
- xx = x.clip(-0.99*max_in, 0.99*max_in)
- return (max_in * numpy.arctanh(xx / max_in)).clip(-max_in, max_in)
- def fisher_z(data):
- """ Fisher''s z-transformation
- For a given dataset :math:`p` bound to :math:`[0.0,1.0]`,we can use Fisher''s z-transformation to normalize it
- in an approximately Gaussian distribution.
- This transformation is computed as follows:
- .. math::
- z_p := \\\\frac{1}{2} \\\\text{ln} \\\\left ( \\\\frac{1+p}{1-p} \\\\right ) = \\\\text{arctanh}(p)
- """
- return np.arctanh(data)
- def test_numpy_method():
- # This type of code is used frequently by PyMC3 users
- x = tt.dmatrix(''x'')
- data = np.random.rand(5, 5)
- x.tag.test_value = data
- for fct in [np.arccos,
- np.arctan, np.ceil, np.cos, np.cosh, np.deg2rad,
- np.exp, np.exp2, np.expm1, np.floor,
- np.log10, np.rad2deg,
- np.sin, np.sinh, np.tan, np.tanh, np.trunc]:
- y = fct(x)
- f = theano.function([x], y)
- utt.assert_allclose(np.nan_to_num(f(data)),
- np.nan_to_num(fct(data)))
- def impl(self, x):
- # If x is an int8 or uint8,numpy.arctanh will compute the result in
- # half-precision (float16),where we want float32.
- x_dtype = str(getattr(x, ''dtype'', ''''))
- if x_dtype in (''int8'', ''uint8''):
- return numpy.arctanh(x, sig=''f'')
- return numpy.arctanh(x)
- def calcAdimCtrl(self,alfa,beta):
- #u = numpy.empty((self.N,self.m))
- Nu = len(alfa)
- u = numpy.empty((Nu,2))
- restrictions = self.restrictions
- alpha_min = restrictions[''alpha_min'']
- alpha_max = restrictions[''alpha_max'']
- beta_min = restrictions[''beta_min'']
- beta_max = restrictions[''beta_max'']
- a1 = .5*(alpha_max + alpha_min)
- a2 = .5*(alpha_max - alpha_min)
- b1 = .5*(beta_max + beta_min)
- b2 = .5*(beta_max - beta_min)
- alfa -= a1
- alfa *= 1.0/a2
- beta -= b1
- beta *= 1.0/b2
- u[:,0] = alfa.copy()
- u[:,1] = beta.copy()
- # Basic saturation
- for j in range(2):
- for k in range(Nu):
- if u[k,j] > 0.99999:
- u[k,j] = 0.99999
- if u[k,j] < -0.99999:
- u[k,j] = -0.99999
- u = numpy.arctanh(u)
- return u
- def arctanh(inp):
- if isinstance(inp, ooarray) and inp.dtype == object:
- return ooarray([arctanh(elem) for elem in inp])
- if not isinstance(inp, oofun):
- return np.arctanh(inp)
- # Todo: move it outside of arctanh deFinition
- def interval(arg_inf, arg_sup):
- raise ''interval for arctanh is unimplemented yet''
- r = oofun(np.arctanh, inp, d = lambda x: FDmisc.Diag(1.0/(1 - x**2)), vectorized = True, interval = interval)
- return r
- def confidence_interval(rho, N):
- """
- Give a 95% confidence interval for a Spearman correlation score,given
- the correlation and the number of cases.
- """
- z = np.arctanh(rho)
- interval = 1.96 / np.sqrt(N - 3)
- low = z - interval
- high = z + interval
- return pd.Series(
- [rho, np.tanh(low), np.tanh(high)],
- index=[''acc'', ''low'', ''high'']
- )
- def test_numpy_ufuncs(self):
- # test ufuncs of numpy 1.9.2. see:
- # http://docs.scipy.org/doc/numpy/reference/ufuncs.html
- # some functions are skipped because it may return different result
- # for unicode input depending on numpy version
- for name, idx in compat.iteritems(self.indices):
- for func in [np.exp,
- np.log1p, np.sin,
- np.arccos,
- np.arcsinh,
- np.rad2deg]:
- if isinstance(idx, pd.tseries.base.DatetimeIndexOpsMixin):
- # raise TypeError or ValueError (Periodindex)
- # Periodindex behavior should be changed in future version
- with tm.assertRaises(Exception):
- func(idx)
- elif isinstance(idx, (Float64Index, Int64Index)):
- # coerces to float (e.g. np.sin)
- result = func(idx)
- exp = Index(func(idx.values), name=idx.name)
- self.assert_index_equal(result, exp)
- self.assertisinstance(result, pd.Float64Index)
- else:
- # raise AttributeError or TypeError
- if len(idx) == 0:
- continue
- else:
- with tm.assertRaises(Exception):
- func(idx)
- for func in [np.isfinite, np.isinf, np.isnan, np.signbit]:
- if isinstance(idx, pd.tseries.base.DatetimeIndexOpsMixin):
- # raise TypeError or ValueError (Periodindex)
- with tm.assertRaises(Exception):
- func(idx)
- elif isinstance(idx, Int64Index)):
- # results in bool array
- result = func(idx)
- exp = func(idx.values)
- self.assertisinstance(result, np.ndarray)
- tm.assertNotisinstance(result, Index)
- else:
- if len(idx) == 0:
- continue
- else:
- with tm.assertRaises(Exception):
- func(idx)

Jupyter 中的 Numpy 在打印时出错(Python 版本 3.8.8):TypeError: 'numpy.ndarray' object is not callable
如何解决Jupyter 中的 Numpy 在打印时出错(Python 版本 3.8.8):TypeError: ''numpy.ndarray'' object is not callable?
晚安, 尝试打印以下内容时,我在 jupyter 中遇到了 numpy 问题,并且得到了一个 错误: 需要注意的是python版本是3.8.8。 我先用 spyder 测试它,它运行正确,它给了我预期的结果
使用 Spyder:
import numpy as np
for i in range (5):
n = np.random.rand ()
print (n)
Results
0.6604903457995978
0.8236300859753154
0.16067650689842816
0.6967868357083673
0.4231597934445466
现在有了 jupyter
import numpy as np
for i in range (5):
n = np.random.rand ()
print (n)
-------------------------------------------------- ------
TypeError Traceback (most recent call last)
<ipython-input-78-0c6a801b3ea9> in <module>
2 for i in range (5):
3 n = np.random.rand ()
----> 4 print (n)
TypeError: ''numpy.ndarray'' object is not callable
感谢您对我如何在 Jupyter 中解决此问题的帮助。
非常感谢您抽出宝贵时间。
阿特,约翰”
解决方法
暂无找到可以解决该程序问题的有效方法,小编努力寻找整理中!
如果你已经找到好的解决方法,欢迎将解决方案带上本链接一起发送给小编。
小编邮箱:dio#foxmail.com (将#修改为@)

numpy.random.random & numpy.ndarray.astype & numpy.arange
今天看到这样一句代码:
xb = np.random.random((nb, d)).astype(''float32'') #创建一个二维随机数矩阵(nb行d列)
xb[:, 0] += np.arange(nb) / 1000. #将矩阵第一列的每个数加上一个值要理解这两句代码需要理解三个函数
1、生成随机数
numpy.random.random(size=None)
size为None时,返回float。
size不为None时,返回numpy.ndarray。例如numpy.random.random((1,2)),返回1行2列的numpy数组
2、对numpy数组中每一个元素进行类型转换
numpy.ndarray.astype(dtype)
返回numpy.ndarray。例如 numpy.array([1, 2, 2.5]).astype(int),返回numpy数组 [1, 2, 2]
3、获取等差数列
numpy.arange([start,]stop,[step,]dtype=None)
功能类似python中自带的range()和numpy中的numpy.linspace
返回numpy数组。例如numpy.arange(3),返回numpy数组[0, 1, 2]

numpy.ravel()/numpy.flatten()/numpy.squeeze()
numpy.ravel(a, order=''C'')
Return a flattened array
numpy.chararray.flatten(order=''C'')
Return a copy of the array collapsed into one dimension
numpy.squeeze(a, axis=None)
Remove single-dimensional entries from the shape of an array.
相同点: 将多维数组 降为 一维数组
不同点:
ravel() 返回的是视图(view),意味着改变元素的值会影响原始数组元素的值;
flatten() 返回的是拷贝,意味着改变元素的值不会影响原始数组;
squeeze()返回的是视图(view),仅仅是将shape中dimension为1的维度去掉;
ravel()示例:
1 import matplotlib.pyplot as plt
2 import numpy as np
3
4 def log_type(name,arr):
5 print("数组{}的大小:{}".format(name,arr.size))
6 print("数组{}的维度:{}".format(name,arr.shape))
7 print("数组{}的维度:{}".format(name,arr.ndim))
8 print("数组{}元素的数据类型:{}".format(name,arr.dtype))
9 #print("数组:{}".format(arr.data))
10
11 a = np.floor(10*np.random.random((3,4)))
12 print(a)
13 log_type(''a'',a)
14
15 a1 = a.ravel()
16 print("a1:{}".format(a1))
17 log_type(''a1'',a1)
18 a1[2] = 100
19
20 print(a)
21 log_type(''a'',a)
flatten()示例
1 import matplotlib.pyplot as plt
2 import numpy as np
3
4 def log_type(name,arr):
5 print("数组{}的大小:{}".format(name,arr.size))
6 print("数组{}的维度:{}".format(name,arr.shape))
7 print("数组{}的维度:{}".format(name,arr.ndim))
8 print("数组{}元素的数据类型:{}".format(name,arr.dtype))
9 #print("数组:{}".format(arr.data))
10
11 a = np.floor(10*np.random.random((3,4)))
12 print(a)
13 log_type(''a'',a)
14
15 a1 = a.flatten()
16 print("修改前a1:{}".format(a1))
17 log_type(''a1'',a1)
18 a1[2] = 100
19 print("修改后a1:{}".format(a1))
20
21 print("a:{}".format(a))
22 log_type(''a'',a)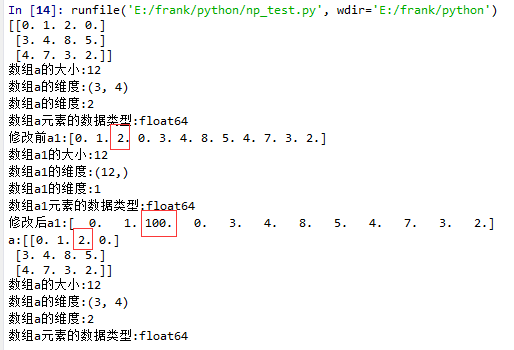
squeeze()示例:
1. 没有single-dimensional entries的情况
1 import matplotlib.pyplot as plt
2 import numpy as np
3
4 def log_type(name,arr):
5 print("数组{}的大小:{}".format(name,arr.size))
6 print("数组{}的维度:{}".format(name,arr.shape))
7 print("数组{}的维度:{}".format(name,arr.ndim))
8 print("数组{}元素的数据类型:{}".format(name,arr.dtype))
9 #print("数组:{}".format(arr.data))
10
11 a = np.floor(10*np.random.random((3,4)))
12 print(a)
13 log_type(''a'',a)
14
15 a1 = a.squeeze()
16 print("修改前a1:{}".format(a1))
17 log_type(''a1'',a1)
18 a1[2] = 100
19 print("修改后a1:{}".format(a1))
20
21 print("a:{}".format(a))
22 log_type(''a'',a)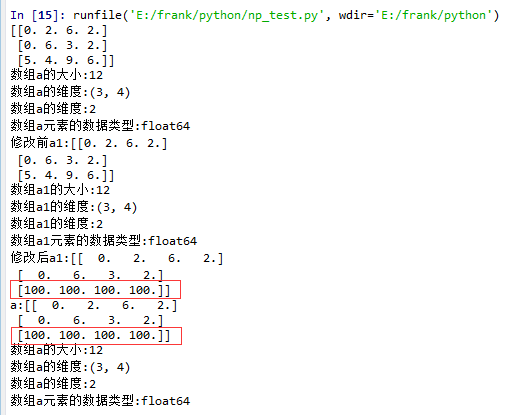
从结果中可以看到,当没有single-dimensional entries时,squeeze()返回额数组对象是一个view,而不是copy。
2. 有single-dimentional entries 的情况
1 import matplotlib.pyplot as plt
2 import numpy as np
3
4 def log_type(name,arr):
5 print("数组{}的大小:{}".format(name,arr.size))
6 print("数组{}的维度:{}".format(name,arr.shape))
7 print("数组{}的维度:{}".format(name,arr.ndim))
8 print("数组{}元素的数据类型:{}".format(name,arr.dtype))
9 #print("数组:{}".format(arr.data))
10
11 a = np.floor(10*np.random.random((1,3,4)))
12 print(a)
13 log_type(''a'',a)
14
15 a1 = a.squeeze()
16 print("修改前a1:{}".format(a1))
17 log_type(''a1'',a1)
18 a1[2] = 100
19 print("修改后a1:{}".format(a1))
20
21 print("a:{}".format(a))
22 log_type(''a'',a)

Numpy:数组创建 numpy.arrray() , numpy.arange()、np.linspace ()、数组基本属性
一、Numpy数组创建
part 1:np.linspace(起始值,终止值,元素总个数
import numpy as np
''''''
numpy中的ndarray数组
''''''
ary = np.array([1, 2, 3, 4, 5])
print(ary)
ary = ary * 10
print(ary)
''''''
ndarray对象的创建
''''''
# 创建二维数组
# np.array([[],[],...])
a = np.array([[1, 2, 3, 4], [5, 6, 7, 8]])
print(a)
# np.arange(起始值, 结束值, 步长(默认1))
b = np.arange(1, 10, 1)
print(b)
print("-------------np.zeros(数组元素个数, dtype=''数组元素类型'')-----")
# 创建一维数组:
c = np.zeros(10)
print(c, ''; c.dtype:'', c.dtype)
# 创建二维数组:
print(np.zeros ((3,4)))
print("----------np.ones(数组元素个数, dtype=''数组元素类型'')--------")
# 创建一维数组:
d = np.ones(10, dtype=''int64'')
print(d, ''; d.dtype:'', d.dtype)
# 创建三维数组:
print(np.ones( (2,3,4), dtype=np.int32 ))
# 打印维度
print(np.ones( (2,3,4), dtype=np.int32 ).ndim) # 返回:3(维)
结果图:

part 2 :np.linspace ( 起始值,终止值,元素总个数)
import numpy as np
a = np.arange( 10, 30, 5 )
b = np.arange( 0, 2, 0.3 )
c = np.arange(12).reshape(4,3)
d = np.random.random((2,3)) # 取-1到1之间的随机数,要求设置为诶2行3列的结构
print(a)
print(b)
print(c)
print(d)
print("-----------------")
from numpy import pi
print(np.linspace( 0, 2*pi, 100 ))
print("-------------np.linspace(起始值,终止值,元素总个数)------------------")
print(np.sin(np.linspace( 0, 2*pi, 100 )))
结果图:

二、Numpy的ndarray对象属性:
数组的结构:array.shape
数组的维度:array.ndim
元素的类型:array.dtype
数组元素的个数:array.size
数组的索引(下标):array[0]
''''''
数组的基本属性
''''''
import numpy as np
print("--------------------案例1:------------------------------")
a = np.arange(15).reshape(3, 5)
print(a)
print(a.shape) # 打印数组结构
print(len(a)) # 打印有多少行
print(a.ndim) # 打印维度
print(a.dtype) # 打印a数组内的元素的数据类型
# print(a.dtype.name)
print(a.size) # 打印数组的总元素个数
print("-------------------案例2:---------------------------")
a = np.array([[1, 2, 3], [4, 5, 6]])
print(a)
# 测试数组的基本属性
print(''a.shape:'', a.shape)
print(''a.size:'', a.size)
print(''len(a):'', len(a))
# a.shape = (6, ) # 此格式可将原数组结构变成1行6列的数据结构
# print(a, ''a.shape:'', a.shape)
# 数组元素的索引
ary = np.arange(1, 28)
ary.shape = (3, 3, 3) # 创建三维数组
print("ary.shape:",ary.shape,"\n",ary )
print("-----------------")
print(''ary[0]:'', ary[0])
print(''ary[0][0]:'', ary[0][0])
print(''ary[0][0][0]:'', ary[0][0][0])
print(''ary[0,0,0]:'', ary[0, 0, 0])
print("-----------------")
# 遍历三维数组:遍历出数组里的每个元素
for i in range(ary.shape[0]):
for j in range(ary.shape[1]):
for k in range(ary.shape[2]):
print(ary[i, j, k], end='' '')
结果图:

关于Python numpy 模块-arctanh() 实例源码和python numpy argmax的介绍现已完结,谢谢您的耐心阅读,如果想了解更多关于Jupyter 中的 Numpy 在打印时出错(Python 版本 3.8.8):TypeError: 'numpy.ndarray' object is not callable、numpy.random.random & numpy.ndarray.astype & numpy.arange、numpy.ravel()/numpy.flatten()/numpy.squeeze()、Numpy:数组创建 numpy.arrray() , numpy.arange()、np.linspace ()、数组基本属性的相关知识,请在本站寻找。
本文标签: