如果您对Pythonnumpy模块-ma()实例源码感兴趣,那么本文将是一篇不错的选择,我们将为您详在本文中,您将会了解到关于Pythonnumpy模块-ma()实例源码的详细内容,我们还将为您解答p
如果您对Python numpy 模块-ma() 实例源码感兴趣,那么本文将是一篇不错的选择,我们将为您详在本文中,您将会了解到关于Python numpy 模块-ma() 实例源码的详细内容,我们还将为您解答python中numpy模块的相关问题,并且为您提供关于Jupyter 中的 Numpy 在打印时出错(Python 版本 3.8.8):TypeError: 'numpy.ndarray' object is not callable、numpy.random.random & numpy.ndarray.astype & numpy.arange、numpy.ravel()/numpy.flatten()/numpy.squeeze()、Numpy:数组创建 numpy.arrray() , numpy.arange()、np.linspace ()、数组基本属性的有价值信息。
本文目录一览:- Python numpy 模块-ma() 实例源码(python中numpy模块)
- Jupyter 中的 Numpy 在打印时出错(Python 版本 3.8.8):TypeError: 'numpy.ndarray' object is not callable
- numpy.random.random & numpy.ndarray.astype & numpy.arange
- numpy.ravel()/numpy.flatten()/numpy.squeeze()
- Numpy:数组创建 numpy.arrray() , numpy.arange()、np.linspace ()、数组基本属性

Python numpy 模块-ma() 实例源码(python中numpy模块)
Python numpy 模块,ma() 实例源码
我们从Python开源项目中,提取了以下44个代码示例,用于说明如何使用numpy.ma()。
- def timer(s, v='''', nloop=500, nrep=3):
- units = ["s", "ms", "µs", "ns"]
- scaling = [1, 1e3, 1e6, 1e9]
- print("%s : %-50s : " % (v, s), end='' '')
- varnames = ["%ss,nm%ss,%sl,nm%sl" % tuple(x*4) for x in ''xyz'']
- setup = ''from __main__ import numpy,ma,%s'' % '',''.join(varnames)
- Timer = timeit.Timer(stmt=s, setup=setup)
- best = min(Timer.repeat(nrep, nloop)) / nloop
- if best > 0.0:
- order = min(-int(numpy.floor(numpy.log10(best)) // 3), 3)
- else:
- order = 3
- print("%d loops,best of %d: %.*g %s per loop" % (nloop, nrep,
- 3,
- best * scaling[order],
- units[order]))
- def timer(s,
- units[order]))
- def fn_getma(fn, bnum=1):
- """Get masked array from input filename
- Parameters
- ----------
- fn : str
- Input filename string
- bnum : int,optional
- Band number
- Returns
- -------
- np.ma.array
- Masked array containing raster values
- """
- #Add check for filename existence
- ds = fn_getds(fn)
- return ds_getma(ds, bnum=bnum)
- #Given input dataset,return a masked array for the input band
- def ds_getma(ds, bnum=1):
- """Get masked array from input GDAL Dataset
- Parameters
- ----------
- ds : gdal.Dataset
- Input GDAL Datset
- bnum : int,optional
- Band number
- Returns
- -------
- np.ma.array
- Masked array containing raster values
- """
- b = ds.GetRasterBand(bnum)
- return b_getma(b)
- #Given input band,return a masked array
- def b_getma(b):
- """Get masked array from input GDAL Band
- Parameters
- ----------
- b : gdal.Band
- Input GDAL Band
- Returns
- -------
- np.ma.array
- Masked array containing raster values
- """
- b_ndv = get_ndv_b(b)
- #bma = np.ma.masked_equal(b.ReadAsArray(),b_ndv)
- #This is more appropriate for float,handles precision issues
- bma = np.ma.masked_values(b.ReadAsArray(), b_ndv)
- return bma
- def timer(s,
- units[order]))
- def timer(s,
- units[order]))
- def timer(s,
- units[order]))
- def test_testPut(self):
- # Test of put
- with suppress_warnings() as sup:
- sup.filter(
- np.ma.core.MaskedArrayFutureWarning,
- "setting an item on a masked array which has a "
- "shared mask will not copy")
- d = arange(5)
- n = [0, 0, 1, 1]
- m = make_mask(n)
- x = array(d, mask=m)
- self.assertTrue(x[3] is masked)
- self.assertTrue(x[4] is masked)
- x[[1, 4]] = [10, 40]
- self.assertTrue(x.mask is not m)
- self.assertTrue(x[3] is masked)
- self.assertTrue(x[4] is not masked)
- self.assertTrue(eq(x, [0, 10, 2, -1, 40]))
- x = array(d, mask=m)
- x.put([0, 2], [-1, 100, 200])
- self.assertTrue(eq(x, 200, 0]))
- self.assertTrue(x[3] is masked)
- self.assertTrue(x[4] is masked)
- def timer(s,
- units[order]))
- def timer(s,
- units[order]))
- def test_testMixedArithmetic(self):
- na = np.array([1])
- ma = array([1])
- self.assertTrue(isinstance(na + ma, MaskedArray))
- self.assertTrue(isinstance(ma + na, MaskedArray))
- def test_testUfuncRegression(self):
- f_invalid_ignore = [
- ''sqrt'', ''arctanh'', ''arcsin'', ''arccos'',
- ''arccosh'', ''log'', ''log10'', ''divide'',
- ''true_divide'', ''floor_divide'', ''remainder'', ''fmod'']
- for f in [''sqrt'', ''exp'', ''conjugate'',
- ''sin'', ''cos'', ''tan'',
- ''arcsin'', ''arctan'',
- ''sinh'', ''cosh'', ''tanh'',
- ''arcsinh'',
- ''arccosh'',
- ''arctanh'',
- ''absolute'', ''fabs'', ''negative'',
- ''floor'', ''ceil'',
- ''logical_not'',
- ''add'', ''subtract'', ''multiply'',
- ''divide'', ''true_divide'',
- ''remainder'', ''fmod'', ''hypot'', ''arctan2'',
- ''equal'', ''not_equal'', ''less_equal'', ''greater_equal'',
- ''less'', ''greater'',
- ''logical_and'', ''logical_or'', ''logical_xor'']:
- try:
- uf = getattr(umath, f)
- except AttributeError:
- uf = getattr(fromnumeric, f)
- mf = getattr(np.ma, f)
- args = self.d[:uf.nin]
- with np.errstate():
- if f in f_invalid_ignore:
- np.seterr(invalid=''ignore'')
- if f in [''arctanh'', ''log10'']:
- np.seterr(divide=''ignore'')
- ur = uf(*args)
- mr = mf(*args)
- self.assertTrue(eq(ur.filled(0), mr.filled(0), f))
- self.assertTrue(eqmask(ur.mask, mr.mask))
- def compare_functions_1v(func,
- xs=xs, nmxs=nmxs, xl=xl, nmxl=nmxl):
- funcname = func.__name__
- print("-"*50)
- print("%s on small arrays" % funcname)
- module, data = "numpy.ma", "nmxs"
- timer("%(module)s.%(funcname)s(%(data)s)" % locals(), v="%11s" % module, nloop=nloop)
- print("%s on large arrays" % funcname)
- module, "nmxl"
- timer("%(module)s.%(funcname)s(%(data)s)" % locals(), nloop=nloop)
- return
- def compare_methods(methodname, args, vars=''x'', test=True,
- xs=xs, nmxl=nmxl):
- print("-"*50)
- print("%s on small arrays" % methodname)
- data, ver = "nm%ss" % vars, ''numpy.ma''
- timer("%(data)s.%(methodname)s(%(args)s)" % locals(), v=ver, nloop=nloop)
- print("%s on large arrays" % methodname)
- data, ver = "nm%sl" % vars, nloop=nloop)
- return
- def test_testMixedArithmetic(self):
- na = np.array([1])
- ma = array([1])
- self.assertTrue(isinstance(na + ma, MaskedArray))
- def test_testUfuncRegression(self):
- f_invalid_ignore = [
- ''sqrt'', mr.mask))
- def compare_functions_1v(func, nloop=nloop)
- return
- def compare_methods(methodname, nloop=nloop)
- return
- def ds_getma_sub(src_ds, bnum=1, scale=None, maxdim=1024.):
- """Load a subsampled array,rather than full resolution
- This is useful when working with large rasters
- Uses buf_xsize and buf_ysize options from GDAL ReadAsArray method.
- Parameters
- ----------
- ds : gdal.Dataset
- Input GDAL Datset
- bnum : int,optional
- Band number
- scale : int,optional
- Scaling factor
- maxdim : int,optional
- Maximum dimension along either axis,in pixels
- Returns
- -------
- np.ma.array
- Masked array containing raster values
- """
- #print src_ds.GetFileList()[0]
- b = src_ds.GetRasterBand(bnum)
- b_ndv = get_ndv_b(b)
- ns, nl = get_sub_dim(src_ds, scale, maxdim)
- #The buf_size parameters determine the final array dimensions
- b_array = b.ReadAsArray(buf_xsize=ns, buf_ysize=nl)
- bma = np.ma.masked_values(b_array, b_ndv)
- return bma
- #Note: need to consolidate with warplib.writeout (takes ds,not ma)
- #Add option to build overviews when writing GTiff
- #Input proj must be WKT
- def replace_ndv(b, new_ndv):
- b_ndv = get_ndv_b(b)
- bma = np.ma.masked_values(b.ReadAsArray(), b_ndv)
- bma.set_fill_value(new_ndv)
- b.WriteArray(bma.filled())
- b.SetNoDataValue(new_ndv)
- return b
- def test_testMixedArithmetic(self):
- na = np.array([1])
- ma = array([1])
- self.assertTrue(isinstance(na + ma, MaskedArray))
- def test_testUfuncRegression(self):
- f_invalid_ignore = [
- ''sqrt'',
- # ''nonzero'',''around'',
- # ''sometrue'',''alltrue'', mr.mask))
- def compare_functions_1v(func, nloop=nloop)
- return
- def compare_methods(methodname, nloop=nloop)
- return
- def test_testMixedArithmetic(self):
- na = np.array([1])
- ma = array([1])
- self.assertTrue(isinstance(na + ma, MaskedArray))
- def test_testUfuncRegression(self):
- f_invalid_ignore = [
- ''sqrt'', mr.mask))
- def compare_functions_1v(func, nloop=nloop)
- return
- def compare_methods(methodname, nloop=nloop)
- return
- def _get_stblock(self, data_input, hnd, mdlt, start_frame=None):
- goodness = False
- if start_frame is None:
- start_frame = random.randint(0, len(data_input[''min_length''])-self.step*(self.nframes-1)-1)
- stblock = numpy.zeros([self.nframes, self.block_size, self.block_size])
- for ii in xrange(self.nframes):
- v = data_input[hnd][mdlt][start_frame + ii * self.step]
- mm = abs(numpy.ma.maximum(v))
- if mm > 0.:
- # normalize to zero mean,unit variance,
- # concatenate in spatio-temporal blocks
- stblock[ii] = self.prenormalize(v)
- goodness = True
- return stblock, goodness
- def test_testMixedArithmetic(self):
- na = np.array([1])
- ma = array([1])
- self.assertTrue(isinstance(na + ma, MaskedArray))
- def test_testUfuncRegression(self):
- f_invalid_ignore = [
- ''sqrt'', mr.mask))
- def compare_functions_1v(func, nloop=nloop)
- return
- def compare_methods(methodname, nloop=nloop)
- return
- def test_testMixedArithmetic(self):
- na = np.array([1])
- ma = array([1])
- self.assertTrue(isinstance(na + ma, MaskedArray))
- def test_testUfuncRegression(self):
- f_invalid_ignore = [
- ''sqrt'', mr.mask))
- def compare_methods(methodname, nloop=nloop)
- return
- def compare_functions_2v(func,
- ys=ys, nmys=nmys,
- xl=xl, nmxl=nmxl,
- yl=yl, nmyl=nmyl):
- funcname = func.__name__
- print("-"*50)
- print("%s on small arrays" % funcname)
- module, "nmxs,nmys"
- timer("%(module)s.%(funcname)s(%(data)s)" % locals(), "nmxl,nmyl"
- timer("%(module)s.%(funcname)s(%(data)s)" % locals(), nloop=nloop)
- return
- def test_testMixedArithmetic(self):
- na = np.array([1])
- ma = array([1])
- self.assertTrue(isinstance(na + ma, MaskedArray))
- def test_testUfuncRegression(self):
- f_invalid_ignore = [
- ''sqrt'', mr.mask))
- def compare_functions_1v(func, nloop=nloop)
- return
- def compare_methods(methodname, nloop=nloop)
- return
- def test_testcopySize(self):
- # Tests of some subtle points of copying and sizing.
- with suppress_warnings() as sup:
- sup.filter(
- np.ma.core.MaskedArrayFutureWarning,
- "setting an item on a masked array which has a "
- "shared mask will not copy")
- n = [0, 0]
- m = make_mask(n)
- m2 = make_mask(m)
- self.assertTrue(m is m2)
- m3 = make_mask(m, copy=1)
- self.assertTrue(m is not m3)
- x1 = np.arange(5)
- y1 = array(x1, mask=m)
- self.assertTrue(y1._data is not x1)
- self.assertTrue(allequal(x1, y1._data))
- self.assertTrue(y1.mask is m)
- y1a = array(y1, copy=0)
- self.assertTrue(y1a.mask is y1.mask)
- y2 = array(x1, mask=m, copy=0)
- self.assertTrue(y2.mask is m)
- self.assertTrue(y2[2] is masked)
- y2[2] = 9
- self.assertTrue(y2[2] is not masked)
- self.assertTrue(y2.mask is not m)
- self.assertTrue(allequal(y2.mask, 0))
- y3 = array(x1 * 1.0, mask=m)
- self.assertTrue(filled(y3).dtype is (x1 * 1.0).dtype)
- x4 = arange(4)
- x4[2] = masked
- y4 = resize(x4, (8,))
- self.assertTrue(eq(concatenate([x4, x4]), y4))
- self.assertTrue(eq(getmask(y4), 0]))
- y5 = repeat(x4, (2, 2), axis=0)
- self.assertTrue(eq(y5, 3, 3]))
- y6 = repeat(x4, y6))
- def composite(reducers, *rasters, normalize=''sum'', nodata=-9999.0, dtype=np.float32):
- ''''''
- NOTE: Uses masked arrays in NumPy and therefore is MUCH slower than the
- `composite2()` function,which is equivalent in output.
- Creates a multi-image (multi-date) composite from input rasters. The
- reducers argument specifies,in the order of the bands (endmembers),how
- to pick a value for that band in each pixel. If None is given,then the
- median value of that band from across the images is used for that pixel
- value. If None is specified as a reducer,the corresponding band(s) will
- be dropped. Combining None reducer(s) with a normalized sum effectively
- subtracts an endmember under the unity constraint. Arguments:
- reducers One of (''min'',''max'',''mean'',''median'',None) for each endmember
- rasters One or more raster files to composite
- normalize True (by default) to normalize results by their sum
- nodata The NoData value (defaults to -9999)
- dtype The data type to coerce in the output array; very important if the desired output is float but NoData value is integer
- ''''''
- shp = rasters[0].shape
- num_non_null_bands = shp[0] - len([b for b in reducers if b is None])
- assert all(map(lambda x: x == shp, [r.shape for r in rasters])), ''Rasters must have the same shape''
- assert len(reducers) == shp[0], ''Must provide a reducer for each band (including None to drop the band)''
- # Swap the sequence of rasters for a sequence of bands,then collapse the X-Y axes
- stack = np.array(rasters).swapaxes(0, 1).reshape(shp[0], len(rasters), shp[-1]*shp[-2])
- # Mask out NoData values
- stack_masked = np.ma.masked_where(stack == nodata, stack)
- # For each band (or endmember)...
- band_arrays = []
- for i in range(shp[0]):
- if reducers[i] in (''min'', ''max'', ''median'', ''mean''):
- band_arrays.append(getattr(np.ma, reducers[i])(stack_masked[i, ...], axis=0))
- # Stack each reduced band (and reshape to multi-band image)
- final_stack = np.ma.vstack(band_arrays).reshape((num_non_null_bands, shp[-2], shp[-1]))
- # Calculate a normalized sum (e.g.,fractions must sum to one)
- if normalize is not None:
- constant = getattr(final_stack, normalize)(axis=0) # The sum across the bands
- constant.set_fill_value(1.0) # NaNs will be divided by 1.0
- constant = np.ma.repeat(constant, num_non_null_bands, axis=0).reshape(final_stack.shape)
- # Divide the values in each band by the normalized sum across the bands
- if num_non_null_bands > 1:
- final_stack = final_stack / constant.swapaxes(0, 1)
- else:
- final_stack = final_stack / constant
- # NOTE: Essential to cast type,e.g.,to float in case first pixel (i.e. top-left) is all NoData of an integer type
- final_stack.set_fill_value(dtype(nodata)) # Fill NoData for NaNs
- return final_stack.filled()

Jupyter 中的 Numpy 在打印时出错(Python 版本 3.8.8):TypeError: 'numpy.ndarray' object is not callable
如何解决Jupyter 中的 Numpy 在打印时出错(Python 版本 3.8.8):TypeError: ''numpy.ndarray'' object is not callable?
晚安, 尝试打印以下内容时,我在 jupyter 中遇到了 numpy 问题,并且得到了一个 错误: 需要注意的是python版本是3.8.8。 我先用 spyder 测试它,它运行正确,它给了我预期的结果
使用 Spyder:
import numpy as np
for i in range (5):
n = np.random.rand ()
print (n)
Results
0.6604903457995978
0.8236300859753154
0.16067650689842816
0.6967868357083673
0.4231597934445466
现在有了 jupyter
import numpy as np
for i in range (5):
n = np.random.rand ()
print (n)
-------------------------------------------------- ------
TypeError Traceback (most recent call last)
<ipython-input-78-0c6a801b3ea9> in <module>
2 for i in range (5):
3 n = np.random.rand ()
----> 4 print (n)
TypeError: ''numpy.ndarray'' object is not callable
感谢您对我如何在 Jupyter 中解决此问题的帮助。
非常感谢您抽出宝贵时间。
阿特,约翰”
解决方法
暂无找到可以解决该程序问题的有效方法,小编努力寻找整理中!
如果你已经找到好的解决方法,欢迎将解决方案带上本链接一起发送给小编。
小编邮箱:dio#foxmail.com (将#修改为@)

numpy.random.random & numpy.ndarray.astype & numpy.arange
今天看到这样一句代码:
xb = np.random.random((nb, d)).astype(''float32'') #创建一个二维随机数矩阵(nb行d列)
xb[:, 0] += np.arange(nb) / 1000. #将矩阵第一列的每个数加上一个值要理解这两句代码需要理解三个函数
1、生成随机数
numpy.random.random(size=None)
size为None时,返回float。
size不为None时,返回numpy.ndarray。例如numpy.random.random((1,2)),返回1行2列的numpy数组
2、对numpy数组中每一个元素进行类型转换
numpy.ndarray.astype(dtype)
返回numpy.ndarray。例如 numpy.array([1, 2, 2.5]).astype(int),返回numpy数组 [1, 2, 2]
3、获取等差数列
numpy.arange([start,]stop,[step,]dtype=None)
功能类似python中自带的range()和numpy中的numpy.linspace
返回numpy数组。例如numpy.arange(3),返回numpy数组[0, 1, 2]
numpy.ravel()/numpy.flatten()/numpy.squeeze()
numpy.ravel(a, order=''C'')
Return a flattened array
numpy.chararray.flatten(order=''C'')
Return a copy of the array collapsed into one dimension
numpy.squeeze(a, axis=None)
Remove single-dimensional entries from the shape of an array.
相同点: 将多维数组 降为 一维数组
不同点:
ravel() 返回的是视图(view),意味着改变元素的值会影响原始数组元素的值;
flatten() 返回的是拷贝,意味着改变元素的值不会影响原始数组;
squeeze()返回的是视图(view),仅仅是将shape中dimension为1的维度去掉;
ravel()示例:
1 import matplotlib.pyplot as plt
2 import numpy as np
3
4 def log_type(name,arr):
5 print("数组{}的大小:{}".format(name,arr.size))
6 print("数组{}的维度:{}".format(name,arr.shape))
7 print("数组{}的维度:{}".format(name,arr.ndim))
8 print("数组{}元素的数据类型:{}".format(name,arr.dtype))
9 #print("数组:{}".format(arr.data))
10
11 a = np.floor(10*np.random.random((3,4)))
12 print(a)
13 log_type(''a'',a)
14
15 a1 = a.ravel()
16 print("a1:{}".format(a1))
17 log_type(''a1'',a1)
18 a1[2] = 100
19
20 print(a)
21 log_type(''a'',a)
flatten()示例
1 import matplotlib.pyplot as plt
2 import numpy as np
3
4 def log_type(name,arr):
5 print("数组{}的大小:{}".format(name,arr.size))
6 print("数组{}的维度:{}".format(name,arr.shape))
7 print("数组{}的维度:{}".format(name,arr.ndim))
8 print("数组{}元素的数据类型:{}".format(name,arr.dtype))
9 #print("数组:{}".format(arr.data))
10
11 a = np.floor(10*np.random.random((3,4)))
12 print(a)
13 log_type(''a'',a)
14
15 a1 = a.flatten()
16 print("修改前a1:{}".format(a1))
17 log_type(''a1'',a1)
18 a1[2] = 100
19 print("修改后a1:{}".format(a1))
20
21 print("a:{}".format(a))
22 log_type(''a'',a)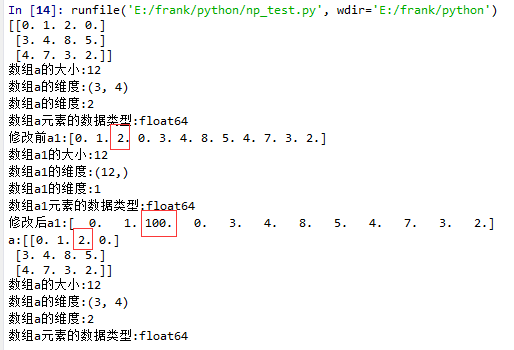
squeeze()示例:
1. 没有single-dimensional entries的情况
1 import matplotlib.pyplot as plt
2 import numpy as np
3
4 def log_type(name,arr):
5 print("数组{}的大小:{}".format(name,arr.size))
6 print("数组{}的维度:{}".format(name,arr.shape))
7 print("数组{}的维度:{}".format(name,arr.ndim))
8 print("数组{}元素的数据类型:{}".format(name,arr.dtype))
9 #print("数组:{}".format(arr.data))
10
11 a = np.floor(10*np.random.random((3,4)))
12 print(a)
13 log_type(''a'',a)
14
15 a1 = a.squeeze()
16 print("修改前a1:{}".format(a1))
17 log_type(''a1'',a1)
18 a1[2] = 100
19 print("修改后a1:{}".format(a1))
20
21 print("a:{}".format(a))
22 log_type(''a'',a)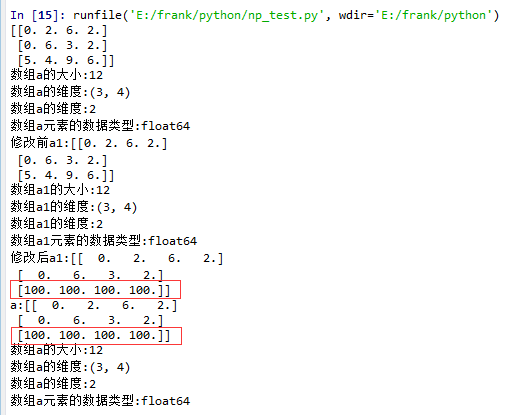
从结果中可以看到,当没有single-dimensional entries时,squeeze()返回额数组对象是一个view,而不是copy。
2. 有single-dimentional entries 的情况
1 import matplotlib.pyplot as plt
2 import numpy as np
3
4 def log_type(name,arr):
5 print("数组{}的大小:{}".format(name,arr.size))
6 print("数组{}的维度:{}".format(name,arr.shape))
7 print("数组{}的维度:{}".format(name,arr.ndim))
8 print("数组{}元素的数据类型:{}".format(name,arr.dtype))
9 #print("数组:{}".format(arr.data))
10
11 a = np.floor(10*np.random.random((1,3,4)))
12 print(a)
13 log_type(''a'',a)
14
15 a1 = a.squeeze()
16 print("修改前a1:{}".format(a1))
17 log_type(''a1'',a1)
18 a1[2] = 100
19 print("修改后a1:{}".format(a1))
20
21 print("a:{}".format(a))
22 log_type(''a'',a)

Numpy:数组创建 numpy.arrray() , numpy.arange()、np.linspace ()、数组基本属性
一、Numpy数组创建
part 1:np.linspace(起始值,终止值,元素总个数
import numpy as np
''''''
numpy中的ndarray数组
''''''
ary = np.array([1, 2, 3, 4, 5])
print(ary)
ary = ary * 10
print(ary)
''''''
ndarray对象的创建
''''''
# 创建二维数组
# np.array([[],[],...])
a = np.array([[1, 2, 3, 4], [5, 6, 7, 8]])
print(a)
# np.arange(起始值, 结束值, 步长(默认1))
b = np.arange(1, 10, 1)
print(b)
print("-------------np.zeros(数组元素个数, dtype=''数组元素类型'')-----")
# 创建一维数组:
c = np.zeros(10)
print(c, ''; c.dtype:'', c.dtype)
# 创建二维数组:
print(np.zeros ((3,4)))
print("----------np.ones(数组元素个数, dtype=''数组元素类型'')--------")
# 创建一维数组:
d = np.ones(10, dtype=''int64'')
print(d, ''; d.dtype:'', d.dtype)
# 创建三维数组:
print(np.ones( (2,3,4), dtype=np.int32 ))
# 打印维度
print(np.ones( (2,3,4), dtype=np.int32 ).ndim) # 返回:3(维)
结果图:

part 2 :np.linspace ( 起始值,终止值,元素总个数)
import numpy as np
a = np.arange( 10, 30, 5 )
b = np.arange( 0, 2, 0.3 )
c = np.arange(12).reshape(4,3)
d = np.random.random((2,3)) # 取-1到1之间的随机数,要求设置为诶2行3列的结构
print(a)
print(b)
print(c)
print(d)
print("-----------------")
from numpy import pi
print(np.linspace( 0, 2*pi, 100 ))
print("-------------np.linspace(起始值,终止值,元素总个数)------------------")
print(np.sin(np.linspace( 0, 2*pi, 100 )))
结果图:

二、Numpy的ndarray对象属性:
数组的结构:array.shape
数组的维度:array.ndim
元素的类型:array.dtype
数组元素的个数:array.size
数组的索引(下标):array[0]
''''''
数组的基本属性
''''''
import numpy as np
print("--------------------案例1:------------------------------")
a = np.arange(15).reshape(3, 5)
print(a)
print(a.shape) # 打印数组结构
print(len(a)) # 打印有多少行
print(a.ndim) # 打印维度
print(a.dtype) # 打印a数组内的元素的数据类型
# print(a.dtype.name)
print(a.size) # 打印数组的总元素个数
print("-------------------案例2:---------------------------")
a = np.array([[1, 2, 3], [4, 5, 6]])
print(a)
# 测试数组的基本属性
print(''a.shape:'', a.shape)
print(''a.size:'', a.size)
print(''len(a):'', len(a))
# a.shape = (6, ) # 此格式可将原数组结构变成1行6列的数据结构
# print(a, ''a.shape:'', a.shape)
# 数组元素的索引
ary = np.arange(1, 28)
ary.shape = (3, 3, 3) # 创建三维数组
print("ary.shape:",ary.shape,"\n",ary )
print("-----------------")
print(''ary[0]:'', ary[0])
print(''ary[0][0]:'', ary[0][0])
print(''ary[0][0][0]:'', ary[0][0][0])
print(''ary[0,0,0]:'', ary[0, 0, 0])
print("-----------------")
# 遍历三维数组:遍历出数组里的每个元素
for i in range(ary.shape[0]):
for j in range(ary.shape[1]):
for k in range(ary.shape[2]):
print(ary[i, j, k], end='' '')
结果图:

今天的关于Python numpy 模块-ma() 实例源码和python中numpy模块的分享已经结束,谢谢您的关注,如果想了解更多关于Jupyter 中的 Numpy 在打印时出错(Python 版本 3.8.8):TypeError: 'numpy.ndarray' object is not callable、numpy.random.random & numpy.ndarray.astype & numpy.arange、numpy.ravel()/numpy.flatten()/numpy.squeeze()、Numpy:数组创建 numpy.arrray() , numpy.arange()、np.linspace ()、数组基本属性的相关知识,请在本站进行查询。
本文标签: