最近很多小伙伴都在问线程“主”org.apache.spark.SparkException中的异常:此JVM中只能运行一个SparkContext和请参阅SPARK-2243这两个问题,那么本篇文章
最近很多小伙伴都在问线程“主” org.apache.spark.SparkException中的异常:此JVM中只能运行一个SparkContext和请参阅SPARK-2243这两个问题,那么本篇文章就来给大家详细解答一下,同时本文还将给你拓展3 pyspark学习---sparkContext概述、Exception in thread "main" org.apache.spark.SparkException: Could not parse Master URL: ''yarn''、org.apache.spark.api.java.JavaSparkContext的实例源码、org.apache.spark.SparkException: A master URL must be set in your configuration等相关知识,下面开始了哦!
本文目录一览:- 线程“主” org.apache.spark.SparkException中的异常:此JVM中只能运行一个SparkContext(请参阅SPARK-2243)
- 3 pyspark学习---sparkContext概述
- Exception in thread "main" org.apache.spark.SparkException: Could not parse Master URL: ''yarn''
- org.apache.spark.api.java.JavaSparkContext的实例源码
- org.apache.spark.SparkException: A master URL must be set in your configuration

线程“主” org.apache.spark.SparkException中的异常:此JVM中只能运行一个SparkContext(请参阅SPARK-2243)
尝试使用cassandra运行spark应用程序时出现错误。
Exception in thread "main" org.apache.spark.SparkException: Only one SparkContext may be running in this JVM (see SPARK-2243).我正在使用Spark版本1.2.0,并且很明显,我在我的应用程序中仅使用了一个Spark上下文。但是每当我尝试添加以下代码进行流式传输时,都会出现此错误。
JavaStreamingContext activitySummaryScheduler = new JavaStreamingContext( sparkConf, new Duration(1000));答案1
小编典典您一次只能拥有一个SparkContext,并且由于StreamingContext中包含一个SparkContext,因此在同一代码中不能有单独的Streaming和Spark
Context。您可以做的是在SparkContext的基础上构建一个StreamingContext,这样,如果您确实需要,就可以访问两者。
使用此构造函数 JavaStreamingContext(sparkContext: JavaSparkContext, batchDuration:Duration)

3 pyspark学习---sparkContext概述
1 Tutorial
Spark本身是由scala语言编写,为了支持py对spark的支持呢就出现了pyspark。它依然可以通过导入Py4j进行RDDS等操作。
2 sparkContext
(1)sparkContext是spark运用的入口点,当我们运行spark的时候,驱动启动同时上下文也开始初始化。
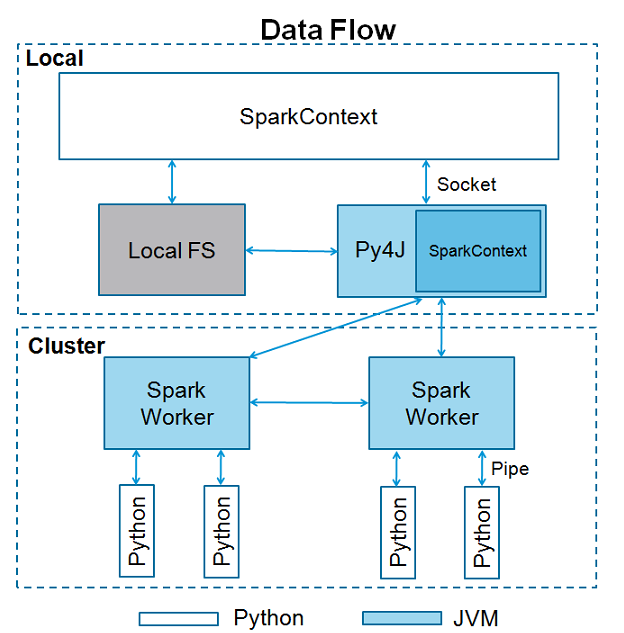
(2)sparkContext使用py4j调用JVM然后创建javaSparkContext,默认为‘sc’,所以如果在shell下就直接用sc.方法就可以。如果你再创建上下文,将会报错cannot run multiple sparkContexts at once哦。结构如下所示
(3)那么一个sparkContext需要哪些内容呢,也就是初始化上下文的时候类有哪些参数呢。
1 class pyspark.SparkContext (
2 master = None,#我们需要连接的集群url
3 appName = None, #工作项目名称
4 sparkHome = None, #spark安装路径
5 pyFiles = None,#一般为处理文件的路径
6 environment = None, #worker节点的环境变量
7 batchSize = 0,
8 serializer = PickleSerializer(), #rdd序列化器
9 conf = None,
10 gateway = None, #要么使用已经存在的JVM要么初始化一个新的JVM
11 jsc = None, #JavaSparkContext实例
12 profiler_cls = <class ''pyspark.profiler.BasicProfiler''>
13 )尝试个例子:在pycharm中使用的哟
1 # coding:utf-8
2 from pyspark import SparkContext, SparkConf
3
4 logFile = "./files/test.txt"
5 sc = SparkContext()
6 logData = sc.textFile(logFile).cache()
7 numA = logData.filter(lambda s: ''a'' in s).count()
8 numB = logData.filter(lambda s: ''a'' in s).count()
9 print "Lines with a: %i, lines with b: %i" % (numA, numB)
加油!

Exception in thread "main" org.apache.spark.SparkException: Could not parse Master URL: ''yarn''

Exception in thread "main" org.apache.spark.SparkException: Could not parse Master URL: ''yarn''
https://stackoverflow.com/questions/41054700/could-not-parse-master-url
缺少 jar 包:
<dependency>
<groupId>org.apache.spark</groupId>
<artifactId>spark-yarn_2.12</artifactId>
<version>${spark.version}</version>
</dependency>

org.apache.spark.api.java.JavaSparkContext的实例源码
public static void main(String[] args) {
if (args.length != 2) {
System.err.println("Usage:");
System.err.println(" SparkWordCount <sourceFile> <targetFile>");
System.exit(1);
}
SparkConf conf = new SparkConf()
.setAppName("Word Count");
JavaSparkContext sc = new JavaSparkContext(conf);
JavaRDD<String> textFile = sc.textFile(args[0]);
JavaRDD<String> words = textFile.flatMap(LineIterator::new);
JavaPairRDD<String,Long> pairs =
words.mapToPair(s -> new Tuple2<>(s,1L));
JavaPairRDD<String,Long> counts =
pairs.reduceByKey((Function2<Long,Long,Long>) (a,b) -> a + b);
System.out.println("Starting task..");
long t = System.currentTimeMillis();
counts.saveAsTextFile(args[1] + "_" + t);
System.out.println("Time=" + (System.currentTimeMillis() - t));
}
/**
* Initializes a Spark connection. Use it afterwards for execution of Spark
* sql queries.
*
* @param appName
* the name of the app that will be used with this Spark
* connection
* @param database
* name of the database that will be used with this Spark
* connection
*/
public Spark(String appName,String database) {
// Todo check what will happen if there is already in use the same app
// name
this.sparkConfiguration = new SparkConf().setAppName(appName);
this.javaContext = new JavaSparkContext(sparkConfiguration);
this.hiveContext = new HiveContext(javaContext);
// Todo check what kind of exception can be thrown here if there is a
// problem with spark connection
this.hiveContext.sql(String.format("CREATE DATABASE %s",database));
// Todo check what kind of exception is thrown if database already
// use the created database
this.hiveContext.sql((String.format("USE %s",database)));
}
/**
* @param sparkContext active Spark Context
* @param trainData training data on which to build a model
* @param hyperParameters ordered list of hyper parameter values to use in building model
* @param candidatePath directory where additional model files can be written
* @return a {@link PMML} representation of a model trained on the given data
*/
@Override
public PMML buildModel(JavaSparkContext sparkContext,JavaRDD<String> trainData,List<?> hyperParameters,Path candidatePath) {
int numClusters = (Integer) hyperParameters.get(0);
Preconditions.checkArgument(numClusters > 1);
log.info("Building KMeans Model with {} clusters",numClusters);
JavaRDD<Vector> trainingData = parsedToVectorRDD(trainData.map(MLFunctions.PARSE_FN));
KMeansModel kMeansModel = KMeans.train(trainingData.rdd(),numClusters,maxIterations,numberOfRuns,initializationStrategy);
return kMeansModelToPMML(kMeansModel,fetchClusterCountsFromModel(trainingData,kMeansModel));
}
public static distributedMatrix GetLU(distributedMatrix A,JavaSparkContext jsc) {
distributedMatrix returnedMatrix;
if( A.getClass() == IndexedRowMatrix.class) {
returnedMatrix = OtherOperations.GetLU_IRW((IndexedRowMatrix) A);
}
else if (A.getClass() == CoordinateMatrix.class) {
returnedMatrix = OtherOperations.GetLU_COORD((CoordinateMatrix) A);
}
else if (A.getClass() == BlockMatrix.class){
// Todo: Implement this operation
//returnedMatrices = OtherOperations.GetLU_BCK((BlockMatrix) A,diagonalInL,diagonalInU,jsc);
returnedMatrix = null;
}
else {
returnedMatrix = null;
}
return returnedMatrix;
}
public static distributedMatrix GetD(distributedMatrix A,boolean inverseValues,JavaSparkContext jsc) {
distributedMatrix returnedMatrix;
if( A.getClass() == IndexedRowMatrix.class) {
returnedMatrix = OtherOperations.GetD_IRW((IndexedRowMatrix) A,inverseValues,jsc);
}
else if (A.getClass() == CoordinateMatrix.class) {
returnedMatrix = OtherOperations.GetD_COORD((CoordinateMatrix) A,jsc);
}
else if (A.getClass() == BlockMatrix.class){
// Todo: Implement this operation
//returnedMatrices = OtherOperations.GetLU_BCK((BlockMatrix) A,jsc);
returnedMatrix = null;
}
else {
returnedMatrix = null;
}
return returnedMatrix;
}
private static void splitFastq(FileStatus fst,String fqPath,String splitDir,int splitlen,JavaSparkContext sc) throws IOException {
Path fqpath = new Path(fqPath);
String fqname = fqpath.getName();
String[] ns = fqname.split("\\.");
//Todo: Handle also compressed files
List<FileSplit> nlif = NLineInputFormat.getSplitsForFile(fst,sc.hadoopConfiguration(),splitlen);
JavaRDD<FileSplit> splitRDD = sc.parallelize(nlif);
splitRDD.foreach( split -> {
FastqRecordReader fqreader = new FastqRecordReader(new Configuration(),split);
writeFastqFile(fqreader,new Configuration(),splitDir + "/split_" + split.getStart() + "." + ns[1]);
});
}
public static void interleaveSplitFastq(FileStatus fst,FileStatus fst2,JavaSparkContext sc) throws IOException {
List<FileSplit> nlif = NLineInputFormat.getSplitsForFile(fst,splitlen);
List<FileSplit> nlif2 = NLineInputFormat.getSplitsForFile(fst2,splitlen);
JavaRDD<FileSplit> splitRDD = sc.parallelize(nlif);
JavaRDD<FileSplit> splitRDD2 = sc.parallelize(nlif2);
JavaPairRDD<FileSplit,FileSplit> zips = splitRDD.zip(splitRDD2);
zips.foreach( splits -> {
Path path = splits._1.getPath();
FastqRecordReader fqreader = new FastqRecordReader(new Configuration(),splits._1);
FastqRecordReader fqreader2 = new FastqRecordReader(new Configuration(),splits._2);
writeInterleavedSplits(fqreader,fqreader2,splitDir+"/"+path.getParent().getName()+"_"+splits._1.getStart()+".fq");
});
}
public static void wordCountJava8( String filename )
{
// Define a configuration to use to interact with Spark
SparkConf conf = new SparkConf().setMaster("local").setAppName("Work Count App");
// Create a Java version of the Spark Context from the configuration
JavaSparkContext sc = new JavaSparkContext(conf);
// Load the input data,which is a text file read from the command line
JavaRDD<String> input = sc.textFile( filename );
// Java 8 with lambdas: split the input string into words
// Todo here a change has happened
JavaRDD<String> words = input.flatMap( s -> Arrays.asList( s.split( " " ) ).iterator() );
// Java 8 with lambdas: transform the collection of words into pairs (word and 1) and then count them
JavaPairRDD<Object,Object> counts = words.mapToPair( t -> new Tuple2( t,1 ) ).reduceByKey( (x,y) -> (int)x + (int)y );
// Save the word count back out to a text file,causing evaluation.
counts.saveAsTextFile( "output" );
}
public static void main(String[] args) throws Exception {
System.out.println(System.getProperty("hadoop.home.dir"));
String inputPath = args[0];
String outputPath = args[1];
FileUtils.deleteQuietly(new File(outputPath));
JavaSparkContext sc = new JavaSparkContext("local","sparkwordcount");
JavaRDD<String> rdd = sc.textFile(inputPath);
JavaPairRDD<String,Integer> counts = rdd
.flatMap(x -> Arrays.asList(x.split(" ")).iterator())
.mapToPair(x -> new Tuple2<String,Integer>((String) x,1))
.reduceByKey((x,y) -> x + y);
counts.saveAsTextFile(outputPath);
sc.close();
}
/**
* loadClickStremFromTxt:Load click stream form txt file
*
* @param clickthroughFile
* txt file
* @param sc
* the spark context
* @return clickstream list in JavaRDD format {@link ClickStream}
*/
public JavaRDD<ClickStream> loadClickStremFromTxt(String clickthroughFile,JavaSparkContext sc) {
return sc.textFile(clickthroughFile).flatMap(new FlatMapFunction<String,ClickStream>() {
/**
*
*/
private static final long serialVersionUID = 1L;
@SuppressWarnings("unchecked")
@Override
public Iterator<ClickStream> call(String line) throws Exception {
List<ClickStream> clickthroughs = (List<ClickStream>) ClickStream.parseFromTextLine(line);
return (Iterator<ClickStream>) clickthroughs;
}
});
}
public SparkDriver(Properties props) {
SparkConf conf = new SparkConf().setAppName(props.getProperty(MudrodConstants.SPARK_APP_NAME,"MudrodSparkApp")).setIfMissing("spark.master",props.getProperty(MudrodConstants.SPARK_MASTER))
.set("spark.hadoop.validateOutputSpecs","false").set("spark.files.overwrite","true");
String esHost = props.getProperty(MudrodConstants.ES_UNICAST_HOSTS);
String esPort = props.getProperty(MudrodConstants.ES_HTTP_PORT);
if (!"".equals(esHost)) {
conf.set("es.nodes",esHost);
}
if (!"".equals(esPort)) {
conf.set("es.port",esPort);
}
conf.set("spark.serializer",KryoSerializer.class.getName());
conf.set("es.batch.size.entries","1500");
sc = new JavaSparkContext(conf);
sqlContext = new sqlContext(sc);
}
private void init() throws IOException {
final ClientConfig config = new ClientConfig();
client = HazelcastClient.newHazelcastClient(config);
final SparkConf conf = new SparkConf()
.set("hazelcast.server.addresses","127.0.0.1:5701")
.set("hazelcast.server.groupName","dev")
.set("hazelcast.server.groupPass","dev-pass")
.set("hazelcast.spark.valueBatchingEnabled","true")
.set("hazelcast.spark.readBatchSize","5000")
.set("hazelcast.spark.writeBatchSize","5000");
sc = new JavaSparkContext("local","appname",conf);
loadHistoricalRaces();
createrandomUsers();
createFutureEvent();
}
/**
*
* @param topKvalueCandidates the topK results per entity,acquired from value similarity
* @param rawTriples1 the rdf triples of the first entity collection
* @param rawTriples2 the rdf triples of the second entity collection
* @param SEParaTOR the delimiter that separates subjects,predicates and objects in the rawTriples1 and rawTriples2 files
* @param entityIds1 the mapping of entity urls to entity ids,as it was used in blocking
* @param entityIds2
* @param MIN_SUPPORT_THRESHOLD the minimum support threshold,below which,relations are discarded from top relations
* @param K the K for topK candidate matches
* @param N the N for topN rdf relations (and neighbors)
* @param jsc the java spark context used to load files and broadcast variables
* @return topK neighbor candidates per entity
*/
public JavaPairRDD<Integer,IntArrayList> run(JavaPairRDD<Integer,Int2FloatLinkedOpenHashMap> topKvalueCandidates,JavaRDD<String> rawTriples1,JavaRDD<String> rawTriples2,String SEParaTOR,JavaRDD<String> entityIds1,JavaRDD<String> entityIds2,float MIN_SUPPORT_THRESHOLD,int K,int N,JavaSparkContext jsc) {
Map<Integer,IntArrayList> inNeighbors = new HashMap<>(new RelationsRank().run(rawTriples1,SEParaTOR,entityIds1,MIN_SUPPORT_THRESHOLD,N,true,jsc));
inNeighbors.putAll(new RelationsRank().run(rawTriples2,entityIds2,false,jsc));
broadcast<Map<Integer,IntArrayList>> inNeighbors_BV = jsc.broadcast(inNeighbors);
//JavaPairRDD<Integer,IntArrayList> topKneighborCandidates = getTopKNeighborSims(topKvalueCandidates,inNeighbors_BV,K);
JavaPairRDD<Integer,IntArrayList> topKneighborCandidates = getTopKNeighborSimsSUM(topKvalueCandidates,K);
return topKneighborCandidates;
}
/**
*
* @param topKvalueCandidates the topK results per entity,Int2FloatLinkedOpenHashMap> run2(JavaPairRDD<Integer,IntArrayList>> inNeighbors_BV = jsc.broadcast(inNeighbors);
JavaPairRDD<Integer,Int2FloatLinkedOpenHashMap> topKneighborCandidates = getTopKNeighborSimsSUMWithscores(topKvalueCandidates,K);
return topKneighborCandidates;
}
/**
*
* @param topKvalueCandidates the topK results per entity,IntArrayList>> inNeighbors_BV = jsc.broadcast(inNeighbors);
//JavaPairRDD<Tuple2<Integer,Integer>,Float> neighborSims = getNeighborSims(topKvalueCandidates,inNeighbors_BV);
//JavaPairRDD<Integer,IntArrayList> topKneighborCandidates = getTopKNeighborSimsOld(neighborSims,K);
return topKneighborCandidates;
}
@Override
public void runUpdate(JavaSparkContext sparkContext,long timestamp,JavaPairRDD<String,String> newData,String> pastData,String modelDirstring,TopicProducer<String,String> modelUpdatetopic) throws IOException {
JavaPairRDD<String,String> allData = pastData == null ? newData : newData.union(pastData);
String modelString;
try {
modelString = new ObjectMapper().writeValueAsstring(countdistinctOtherWords(allData));
} catch (JsonProcessingException jpe) {
throw new IOException(jpe);
}
modelUpdatetopic.send("MODEL",modelString);
}
@Before
public void setUp() {
System.setProperty("hadoop.home.dir","C:\\Users\\VASILIS\\Documents\\hadoop_home"); //only for local mode
spark = SparkSession.builder()
.appName("test")
.config("spark.sql.warehouse.dir","/file:/tmp")
.config("spark.executor.instances",1)
.config("spark.executor.cores",1)
.config("spark.executor.memory","1G")
.config("spark.driver.maxResultSize","1g")
.config("spark.master","local")
.getorCreate();
jsc = JavaSparkContext.fromSparkContext(spark.sparkContext());
}
@Before
public void setUp() {
System.setProperty("hadoop.home.dir","local")
.getorCreate();
jsc = JavaSparkContext.fromSparkContext(spark.sparkContext());
}
@BeforeClass
public static void createContext() throws IOException {
Configuration hdfsConfig = HDfsutils.getConfiguration();
SparkConf config = new SparkConf();
config.setMaster("local[*]");
config.setAppName("my JUnit running Spark");
sc = new JavaSparkContext(config);
fileSystem = FileSystem.get(hdfsConfig);
sqlContext = new sqlContext(sc);
engine = new ParquetRepartEngine(fileSystem,sqlContext);
}
private static MatrixFactorizationModel pmmlToMFModel(JavaSparkContext sparkContext,PMML pmml,Path modelParentPath,broadcast<Map<String,Integer>> bUserIDToIndex,Integer>> bItemIDToIndex) {
String xPathString = AppPMMLUtils.getExtensionValue(pmml,"X");
String yPathString = AppPMMLUtils.getExtensionValue(pmml,"Y");
JavaPairRDD<String,float[]> userRDD = readFeaturesRDD(sparkContext,new Path(modelParentPath,xPathString));
JavaPairRDD<String,float[]> productRDD = readFeaturesRDD(sparkContext,yPathString));
int rank = userRDD.first()._2().length;
return new MatrixFactorizationModel(
rank,readAndConvertFeatureRDD(userRDD,bUserIDToIndex),readAndConvertFeatureRDD(productRDD,bItemIDToIndex));
}
public static void main( String[] args )
{
Dataset<Row> mutations = DataProvider.getMutationsToStructures();
List<String> pdbIds = mutations.select(col("pdbId"))
.distinct().toJavaRDD().map(t -> t.getString(0)).collect();
List<Row> broadcasted = mutations.select("pdbId","chainId","pdbAtomPos").collectAsList();
SaprkUtils.stopSparkSession();
JavaSparkContext sc = SaprkUtils.getSparkContext();
broadcast<List<Row>> bcmut = sc.broadcast(broadcasted);
MmtfReader//.readSequenceFile("/pdb/2017/full",pdbIds,sc)
.downloadMmtfFiles(Arrays.asList("5IRC"),sc)
.flatMapToPair(new StructuretopolymerChains())
.flatMapToPair(new AddResiduetoKey(bcmut))
.mapValues(new StructuretoBioJava())
.mapToPair(new FilterResidue())
.filter(t -> t._2!=null).keys()
.map(t -> t.replace(".",","))
.saveAsTextFile("/Users/yana/git/mutantpdb/src/main/resources/pdb_residues");
sc.close();
}
public static distributedMatrix[] GetLU(distributedMatrix A,boolean diagonalInL,boolean diagonalInU,JavaSparkContext jsc) {
if((diagonalInL && diagonalInU) || (!diagonalInL && !diagonalInU)) {
LOG.error("Diagonal values must be in either upper or lower matrices");
System.exit(-1);
}
distributedMatrix[] returnedMatrices;
if( A.getClass() == IndexedRowMatrix.class) {
returnedMatrices = OtherOperations.GetLU_IRW((IndexedRowMatrix) A,jsc);
}
else if (A.getClass() == CoordinateMatrix.class) {
returnedMatrices = OtherOperations.GetLU_COORD((CoordinateMatrix) A,jsc);
returnedMatrices = null;
}
else {
returnedMatrices = null;
}
return returnedMatrices;
}
public static DenseVector DGEMV(double alpha,distributedMatrix A,DenseVector x,double beta,DenseVector y,JavaSparkContext jsc){
// First form y := beta*y.
if (beta != 1.0) {
if (beta == 0.0) {
y = Vectors.zeros(y.size()).toDense();
}
else {
BLAS.scal(beta,y);
}
}
if (alpha == 0.0) {
return y;
}
DenseVector tmpVector = Vectors.zeros(y.size()).toDense();
// Form y := alpha*A*x + y.
// Case of IndexedRowMatrix
if( A.getClass() == IndexedRowMatrix.class) {
tmpVector = L2.DGEMV_IRW((IndexedRowMatrix) A,alpha,x,jsc);
}
else if (A.getClass() == CoordinateMatrix.class) {
tmpVector = L2.DGEMV_COORD((CoordinateMatrix) A,jsc);
}
else if (A.getClass() == BlockMatrix.class){
tmpVector = L2.DGEMV_BCK((BlockMatrix) A,jsc);
}
else {
tmpVector = null;
}
BLAS.axpy(1.0,tmpVector,y);
return y;
}
private static DenseVector DGEMV_COORD(CoordinateMatrix matrix,double alpha,DenseVector vector,JavaSparkContext jsc) {
JavaRDD<MatrixEntry> items = matrix.entries().toJavaRDD();
DenseVector result = items.mapPartitions(new MatrixEntriesMultiplication(vector,alpha))
.reduce(new MatrixEntriesMultiplicationReducer());
return result;
}
public static void main(String[] args) {
SparkConf sc = new SparkConf().setAppName("POC-Batch");
try(JavaSparkContext jsc = new JavaSparkContext(sc)) {
JavaRDD<ExampleXML> records = jsc.wholeTextFiles("input/")
.map(t -> t._2())
.map(new ParseXML());
System.out.printf("Amount of XMLs: %d\n",records.count());
}
}
public DecisionTreeValidationSummary validateDecisionTreeAthenaFeatures(JavaSparkContext sc,FeatureConstraint featureConstraint,AthenaMLFeatureConfiguration athenaMLFeatureConfiguration,DecisionTreeDetectionModel decisionTreeDetectionModel,Indexing indexing,Marking marking) {
long start = System.nanoTime(); // <-- start
JavaPairRDD<Object,BSONObject> mongoRDD;
mongoRDD = sc.newAPIHadoopRDD(
mongodbConfig,// Configuration
MongoInputFormat.class,// InputFormat: read from a live cluster.
Object.class,// Key class
BSONObject.class // Value class
);
DecisionTreeDetectionAlgorithm decisionTreeDetectionAlgorithm = (DecisionTreeDetectionAlgorithm) decisionTreeDetectionModel.getDetectionAlgorithm();
DecisionTreeValidationSummary decisionTreeValidationSummary =
new DecisionTreeValidationSummary(sc.sc(),decisionTreeDetectionAlgorithm.getNumClasses(),indexing,marking);
DecisionTreedistJob decisionTreedistJob = new DecisionTreedistJob();
decisionTreedistJob.validate(mongoRDD,athenaMLFeatureConfiguration,decisionTreeDetectionModel,decisionTreeValidationSummary);
long end = System.nanoTime(); // <-- start
long time = end - start;
decisionTreeValidationSummary.setTotalValidationTime(time);
return decisionTreeValidationSummary;
}
public GaussianMixtureValidationSummary validateGaussianMixtureAthenaFeatures(JavaSparkContext sc,GaussianMixtureDetectionModel gaussianMixtureDetectionModel,// Key class
BSONObject.class // Value class
);
GaussianMixtureDetectionAlgorithm gaussianMixtureDetectionAlgorithm = (GaussianMixtureDetectionAlgorithm) gaussianMixtureDetectionModel.getDetectionAlgorithm();
GaussianMixtureValidationSummary gaussianMixtureValidationSummary =
new GaussianMixtureValidationSummary(sc.sc(),gaussianMixtureDetectionAlgorithm.getK(),marking);
GaussianMixturedistJob gaussianMixturedistJob = new GaussianMixturedistJob();
gaussianMixturedistJob.validate(mongoRDD,gaussianMixtureDetectionModel,gaussianMixtureValidationSummary);
long end = System.nanoTime(); // <-- start
long time = end - start;
gaussianMixtureValidationSummary.setTotalValidationTime(time);
return gaussianMixtureValidationSummary;
}
public static void main( String[] args ){
String inputFile = "data/dummy.txt";
SparkConf configuration = new SparkConf().setMaster("local[4]").setAppName("My App");
JavaSparkContext sparkContext = new JavaSparkContext(configuration);
JavaRDD<String> logData = sparkContext.textFile(inputFile).cache();
long numberA = logData.filter(new Function<String,Boolean>(){
private static final long serialVersionUID = 1L;
public Boolean call(String s){
return s.length() == 0;
}
}).count();
sparkContext.close();
System.out.println("Empty Lines: " + numberA);
}
public LassovalidationSummary validateLassoAthenaFeatures(JavaSparkContext sc,LassoDetectionModel lassoDetectionModel,// Key class
BSONObject.class // Value class
);
LassoDetectionAlgorithm lassoDetectionAlgorithm =
(LassoDetectionAlgorithm) lassoDetectionModel.getDetectionAlgorithm();
LassovalidationSummary lassovalidationSummary = new LassovalidationSummary();
lassovalidationSummary.setLassoDetectionAlgorithm(lassoDetectionAlgorithm);
LassodistJob lassodistJob = new LassodistJob();
lassodistJob.validate(mongoRDD,lassoDetectionModel,lassovalidationSummary);
long end = System.nanoTime(); // <-- start
long time = end - start;
lassovalidationSummary.setValidationTime(time);
return lassovalidationSummary;
}
public RidgeRegressionValidationSummary validateRidgeRegressionAthenaFeatures(JavaSparkContext sc,RidgeRegressionDetectionModel ridgeRegressionDetectionModel,// Key class
BSONObject.class // Value class
);
RidgeRegressionDetectionAlgorithm ridgeRegressionDetectionAlgorithm =
(RidgeRegressionDetectionAlgorithm) ridgeRegressionDetectionModel.getDetectionAlgorithm();
RidgeRegressionValidationSummary ridgeRegressionValidationSummary = new RidgeRegressionValidationSummary();
ridgeRegressionValidationSummary.setRidgeRegressionDetectionAlgorithm(ridgeRegressionDetectionAlgorithm);
RidgeRegressiondistJob ridgeRegressiondistJob = new RidgeRegressiondistJob();
ridgeRegressiondistJob.validate(mongoRDD,ridgeRegressionDetectionModel,ridgeRegressionValidationSummary);
long end = System.nanoTime(); // <-- start
long time = end - start;
ridgeRegressionValidationSummary.setValidationTime(time);
return ridgeRegressionValidationSummary;
}
private static void splitFastq(FileStatus fst,JavaSparkContext sc) throws IOException {
Path fqpath = new Path(fqPath);
String fqname = fqpath.getName();
String[] ns = fqname.split("\\.");
List<FileSplit> nlif = NLineInputFormat.getSplitsForFile(fst,splitDir + "/split_" + split.getStart() + "." + ns[1]);
});
}
public static void interleaveSplitFastq(FileStatus fst,splits._2);
writeInterleavedSplits(fqreader,splitDir+"/"+path.getParent().getName()+"_"+splits._1.getStart()+".fq");
});
}
public static SparkUtils getInstance() {
SparkUtils result = instance;
if (result == null) {
synchronized (SparkUtils.class) {
result = instance;
if (result == null) {
instance = result = new SparkUtils();
SparkConf sparkConf = new SparkConf().setMaster("local").setAppName("Explainer");
jsc = new JavaSparkContext(sparkConf);
sqlContext = new sqlContext(jsc);
}
}
}
return result;
}
@Override
public void publishAdditionalModelData(JavaSparkContext sparkContext,JavaRDD<String> newData,JavaRDD<String> pastData,String> modelUpdatetopic) {
// Send item updates first,before users. That way,user-based endpoints like /recommend
// may take longer to not return 404,but when they do,the result will be more complete.
log.info("Sending item / Y data as model updates");
String yPathString = AppPMMLUtils.getExtensionValue(pmml,yPathString));
String updatebroker = modelUpdatetopic.getUpdatebroker();
String topic = modelUpdatetopic.getTopic();
// For Now,there is no use in sending kNown users for each item
productRDD.foreachPartition(new EnqueueFeatureVecsFn("Y",updatebroker,topic));
log.info("Sending user / X data as model updates");
String xPathString = AppPMMLUtils.getExtensionValue(pmml,"X");
JavaPairRDD<String,xPathString));
if (noKNownItems) {
userRDD.foreachPartition(new EnqueueFeatureVecsFn("X",topic));
} else {
log.info("Sending kNown item data with model updates");
JavaRDD<String[]> allData =
(pastData == null ? newData : newData.union(pastData)).map(MLFunctions.PARSE_FN);
JavaPairRDD<String,Collection<String>> kNownItems = kNownsRDD(allData,true);
userRDD.join(kNownItems).foreachPartition(
new EnqueueFeatureVecsAndKNownItemsFn("X",topic));
}
}
public NaiveBayesValidationSummary validateNaiveBayesAthenaFeatures(JavaSparkContext sc,NaiveBayesDetectionModel naiveBayesDetectionModel,// Key class
BSONObject.class // Value class
);
NaiveBayesDetectionAlgorithm naiveBayesDetectionAlgorithm = (NaiveBayesDetectionAlgorithm) naiveBayesDetectionModel.getDetectionAlgorithm();
NaiveBayesValidationSummary naiveBayesValidationSummary =
new NaiveBayesValidationSummary(sc.sc(),naiveBayesDetectionAlgorithm.getNumClasses(),marking);
NaiveBayesdistJob naiveBayesdistJob = new NaiveBayesdistJob();
naiveBayesdistJob.validate(mongoRDD,naiveBayesDetectionModel,naiveBayesValidationSummary);
long end = System.nanoTime(); // <-- start
long time = end - start;
naiveBayesValidationSummary.setTotalValidationTime(time);
return naiveBayesValidationSummary;
}
public GradientBoostedTreesValidationSummary validateGradientBoostedTreesAthenaFeatures(JavaSparkContext sc,GradientBoostedTreesDetectionModel gradientBoostedTreesDetectionModel,// Key class
BSONObject.class // Value class
);
GradientBoostedTreesDetectionAlgorithm gradientBoostedTreesDetectionAlgorithm =
(GradientBoostedTreesDetectionAlgorithm) gradientBoostedTreesDetectionModel.getDetectionAlgorithm();
GradientBoostedTreesValidationSummary gradientBoostedTreesValidationSummary =
new GradientBoostedTreesValidationSummary(sc.sc(),gradientBoostedTreesDetectionAlgorithm.getNumClasses(),marking);
GradientBoostedTreesdistJob gradientBoostedTreesdistJob = new GradientBoostedTreesdistJob();
gradientBoostedTreesdistJob.validate(mongoRDD,gradientBoostedTreesDetectionModel,gradientBoostedTreesValidationSummary);
long end = System.nanoTime(); // <-- start
long time = end - start;
gradientBoostedTreesValidationSummary.setTotalValidationTime(time);
return gradientBoostedTreesValidationSummary;
}
@Provides
CassandraIo<Rawrating> providesCassandraratingIO(JavaSparkContext sparkContext) {
if (ratingCassandraIo != null) {
return ratingCassandraIo;
}
ratingCassandraIo = new CassandraIo<>(Rawrating.class,"dev","ratings");
ratingCassandraIo.setSparkContext(sparkContext);
return ratingCassandraIo;
}
public static void main(String[] args) throws ParseException {
final Validator validator = new Validator(args);
ValidatorParameters params = validator.getParameters();
validator.setDoPrintInProcessRecord(false);
logger.info("Input file is " + params.getArgs());
SparkConf conf = new SparkConf().setAppName("marcCompletenessCount");
JavaSparkContext context = new JavaSparkContext(conf);
System.err.println(validator.getParameters().formatParameters());
JavaRDD<String> inputFile = context.textFile(validator.getParameters().getArgs()[0]);
JavaRDD<String> baseCountsRDD = inputFile
.flatMap(content -> {
marcReader reader = Readmarc.getmarcStringReader(content);
Record marc4jRecord = reader.next();
marcRecord marcRecord = marcFactory.createFrommarc4j(
marc4jRecord,params.getDefaultrecordtype(),params.getmarcVersion(),params.fixalephseq());
validator.processRecord(marcRecord,1);
return ValidationErrorFormatter
.formatForSummary(marcRecord.getValidationErrors(),params.getFormat())
.iterator();
}
);
baseCountsRDD.saveAsTextFile(validator.getParameters().getFileName());
}
public LinearRegressionValidationSummary validateLinearRegressionAthenaFeatures(JavaSparkContext sc,LinearRegressionDetectionModel linearRegressionDetectionModel,// Key class
BSONObject.class // Value class
);
LinearRegressionDetectionAlgorithm linearRegressionDetectionAlgorithm =
(LinearRegressionDetectionAlgorithm) linearRegressionDetectionModel.getDetectionAlgorithm();
LinearRegressionValidationSummary linearRegressionValidationSummary =
new LinearRegressionValidationSummary();
linearRegressionValidationSummary.setLinearRegressionDetectionAlgorithm(linearRegressionDetectionAlgorithm);
LinearRegressiondistJob linearRegressiondistJob = new LinearRegressiondistJob();
linearRegressiondistJob.validate(mongoRDD,linearRegressionDetectionModel,linearRegressionValidationSummary);
long end = System.nanoTime(); // <-- start
long time = end - start;
linearRegressionValidationSummary.setValidationTime(time);
return linearRegressionValidationSummary;
}
/**
* Parse RDF file from resources folder
* @param sc spark context to use for parsing
* @param fileName name of the file to parse
* @return RDD of quads from the requested file
*/
public static JavaRDD<Quad> getQuadsRDD(JavaSparkContext sc,String fileName) {
QuadParser parser = new elephasQuadParser(
new QuadParserConfig()
.setBatchSize(2),sc
);
String path = TestUtils.getDatasetPath(fileName);
return parser.parseQuads(path);
}

org.apache.spark.SparkException: A master URL must be set in your configuration
执行 spark examples 报错:
Using Spark''s default log4j profile: org/apache/spark/log4j-defaults.properties
18/05/25 11:09:45 INFO SparkContext: Running Spark version 2.1.1
18/05/25 11:09:45 WARN NativeCodeLoader: Unable to load native-hadoop library for your platform... using builtin-java classes where applicable
18/05/25 11:09:45 ERROR SparkContext: Error initializing SparkContext.
org.apache.spark.SparkException: A master URL must be set in your configuration
at org.apache.spark.SparkContext.<init>(SparkContext.scala:379)
at org.apache.spark.SparkContext$.getOrCreate(SparkContext.scala:2320)
at org.apache.spark.sql.SparkSession$Builder$$anonfun$6.apply(SparkSession.scala:868)
at org.apache.spark.sql.SparkSession$Builder$$anonfun$6.apply(SparkSession.scala:860)
at scala.Option.getOrElse(Option.scala:121)
at org.apache.spark.sql.SparkSession$Builder.getOrCreate(SparkSession.scala:860)
at org.apache.spark.examples.graphx.AggregateMessagesExample$.main(AggregateMessagesExample.scala:42)
at org.apache.spark.examples.graphx.AggregateMessagesExample.main(AggregateMessagesExample.scala)
18/05/25 11:09:45 INFO SparkContext: Successfully stopped SparkContext
Exception in thread "main" org.apache.spark.SparkException: A master URL must be set in your configuration
at org.apache.spark.SparkContext.<init>(SparkContext.scala:379)
at org.apache.spark.SparkContext$.getOrCreate(SparkContext.scala:2320)
at org.apache.spark.sql.SparkSession$Builder$$anonfun$6.apply(SparkSession.scala:868)
at org.apache.spark.sql.SparkSession$Builder$$anonfun$6.apply(SparkSession.scala:860)
at scala.Option.getOrElse(Option.scala:121)
at org.apache.spark.sql.SparkSession$Builder.getOrCreate(SparkSession.scala:860)
at org.apache.spark.examples.graphx.AggregateMessagesExample$.main(AggregateMessagesExample.scala:42)
at org.apache.spark.examples.graphx.AggregateMessagesExample.main(AggregateMessagesExample.scala)
Process finished with exit code 1解决方法:在启动 vm 里添加参数
-Dspark.master=local
关于线程“主” org.apache.spark.SparkException中的异常:此JVM中只能运行一个SparkContext和请参阅SPARK-2243的介绍现已完结,谢谢您的耐心阅读,如果想了解更多关于3 pyspark学习---sparkContext概述、Exception in thread "main" org.apache.spark.SparkException: Could not parse Master URL: ''yarn''、org.apache.spark.api.java.JavaSparkContext的实例源码、org.apache.spark.SparkException: A master URL must be set in your configuration的相关知识,请在本站寻找。
本文标签: